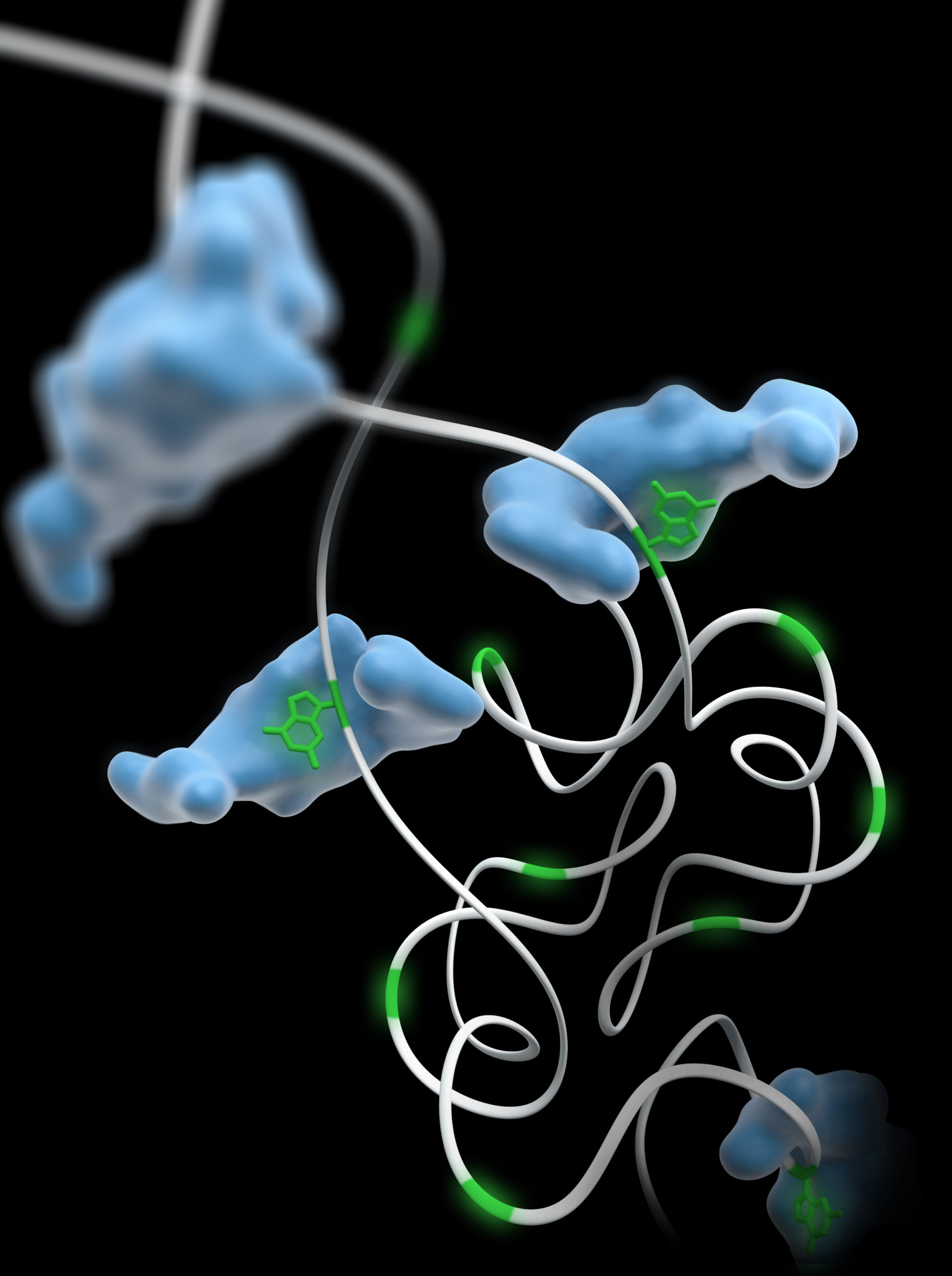
Researchers at the University of North Carolina at Chapel Hill have discovered how helper molecules, or chaperones, aid in the RNA folding process, resolving a fundamental conundrum about how these important biological molecules work.
The research, published online in the March 7 issue of Science, was led by graduate student Jake Grohman in the chemistry department laboratory of Kevin Weeks, Kenan Distinguished Professor in UNC’s College of Arts and Sciences and a member of the UNC Lineberger Comprehensive Cancer Center.
Many important biological molecules only work properly in cells after they have assembled into the correct three-dimensional shape. Single-stranded, ribbon-like RNA molecules are able to fold into intricate patterns and structures, a complex process. The role that RNA chaperones play in this process had not been previously well understood, noted Weeks.
“For example, these chaperones can help some RNAs interact faster, can help other RNAs change conformation, and can work simultaneously across large RNAs. It was not clear how these simple chaperones could have such molecular Mary Poppins-like qualities,” Weeks said.
RNA structures contain many base pairs, mostly between nucleotides (building blocks) called guanosine and cytosine, and between adenosine and uridine. The guanosine-cytosine pairs are stabilized by three bonds, while adenosine-uridine pairs have only two bonds. Sometimes, due to the three bonds, the stronger guanosine-cytosine pairs get “stuck” during the folding process.
“We discovered that a big part of what RNA chaperones do is to weaken the three-bond guanosine-cytosine pairs,” Weeks said. “Thus, RNA chaperones function like molecular lubricants to smooth out the rough spots in forming the correct sets of interactions that allow RNA to function in cells.”
The researchers performed their study using a model retrovirus, simpler than — but similar in many respects to — HIV, using novel chemical microscopes developed in Weeks’ laboratory. In 2009, Weeks and UNC researchers decoded the structure of an entire HIV genome.
“Many RNA viruses require specialized RNA chaperones to replicate and, over the long term, understanding how to disrupt viral chaperones might be a useful approach for developing anti-viral interventions,” Weeks said.
The study involved collaborations between the UNC departments of chemistry, biology, biophysics and biochemistry, and with the National Cancer Institute and St. Louis University.
Additional UNC researchers include Colin Lickwar and Jason Lieb in the department of biology, and Brian Bower in the department of genetics in the School of Medicine. Lieb also is director of the Carolina Center for Genome Sciences and a member of UNC Lineberger.